MYC promotes POLR3G gene expression, shaping Pol III identity and downstream transcription activities associated with cell proliferation.
Urbana, Ill. – Researchers at the University of Illinois Urbana-Champaign and Stanford University School of Medicine have identified a link between RNA polymerase III subunit composition and transcription, an advancement that has potential implications for future cancer research. A switch in one individual subunit, which gives rise to alternate Pol III “identities”, affects the ability of Pol III to express specific small RNAs with downstream roles in cancer growth and metastasis.
Pol III is a large, multi-subunit complex that transcribes a specific subset of genes that don’t encode for proteins, but rather for small RNAs that play critical roles in biology.
“Pol III is often thought of as a ‘housekeeper’ that expresses genes that are critical for cell growth and proliferation” said first author Kevin Van Bortle, CCIL scientist and professor of cell and developmental biology. “It’s the synthesizer of many critical classes of small RNA.”
Van Bortle and colleagues built an “unprecedented” genomic map of RNA polymerase III activity in the nucleus by mapping nearly half of the subunits of Pol III in an experimental approach known as chromatin immunoprecipitation (ChIP), which uses antibodies to pull down specific proteins. The findings were recently published in Nature Communications. Van Bortle says his group wanted to know the full extent of Pol III transcription and how its activity changes during differentiation and in cancer.
“Accurately profiling changes in transcription presents certain challenges, and for Pol III, there are added challenges related to mapping sequence reads to multicopy genes” Van Bortle says. “The approach of ChIP, which pulls down large DNA fragments, helps us map and focus on genes that are actually expressed when we might otherwise mismap RNA reads to genes that are not active.”

Kevin Van Bortle.
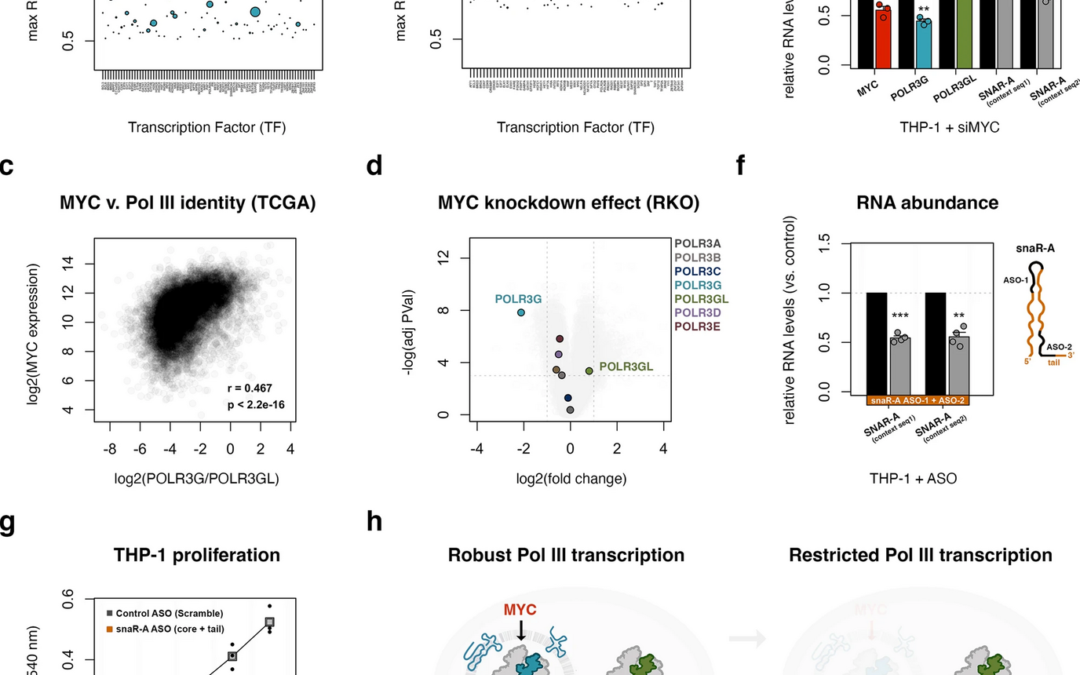
Van Bortle says Pol III is unique compared to other RNA polymerase complexes because it turns off one subunit, POLR3G, and switches it with a different protein, POLR3GL, during development. The genomic map of Pol III activity helped identify a subset of Pol III transcribed-genes that are strongly enhanced or even dependent on subunit POLR3G. Lab researchers went on to demonstrate that the POLR3G-dependent small RNA actively promote proliferation in cancer cells, linking Pol III composition “identity” to transcription as well as downstream growth.
The team also investigated the connection between Pol III identity and survival outcomes in multiple cancers. Van Bortle says POLR3G reemerges in many, but not all, cancer contexts, and is linked to poor survival outcomes in those specific subsets, including lung, bladder, and kidney cancers. He’s interested in the biological implication his research will have on treating cancer. The lab’s next steps include determining whether the reemergence of POLR3G is important in specific cancers, therefore translating to the development of drugs targeting the subunit.
“It’s exciting to follow this path of both understanding the basic science and making discoveries that could have an impact on human health… In a few years from now, we’ll be interested in developing and testing drug libraries for inhibiting both Pol III generally and this subunit, specifically with the hope that there would be long-term therapeutic potential.”
– Written by Jennifer Lask.
The paper, “A cancer-associated RNA polymerase III identity drives robust transcription and expression of snaR-A noncoding RNA,” is available online. DOI: 10.1038/s41467-022-30323-6