Image of Brian Cunningham
Urbana, Ill. – Cancer Center at Illinois and Mount Sinai researchers have developed a low-cost, portable, point-of-care technology capable of diagnosing early-stage liver cancer within 30 minutes.
The study, led by Brian Cunningham, Cancer Center at Illinois program leader and professor of electrical and computer engineering, and Carl R. Woese Institute for Genomic Biology (IGB) postdoctoral fellow Bin Zhao, uses a toaster-sized device comprised of a red LED light, microscope objective, and webcam to detect gold nanoparticles (AuNPs) that attach to target RNA.
Researchers from the Mount Sinai group had previously identified three snRNA biomarkers that were relevant for diagnosing liver cancer, and Cunningham’s group developed the technology, a photonic resonator absorption microscopy (PRAM) assay, for detecting low levels of snRNA. This study was the first clinical collaboration testing the technology with human specimens.
“The detection instrument, PRAM, is very inexpensive, small, and has about $7,000 worth of components including the computer, which makes it ideal for point-of-care diagnostic tests for genomic mutations in the clinic,” said Cunningham.
This testing method requires only a few drops of the patient’s blood, is sensitive enough to detect microRNAs (miRNAs) in serum, and is much faster than PCR, the predominant method of detecting snRNAs.
PCR can be very sensitive but requires a laboratory environment where researchers can amplify the molecules with enzymes and multiple thermal cycles that necessitate accurate temperatures in each step of the process.
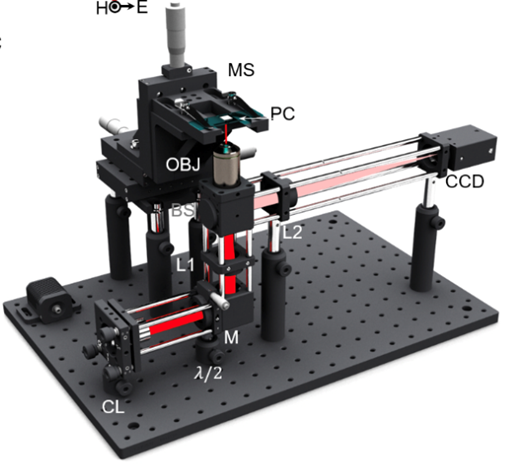
PRAM instrument
“The process doesn’t happen all at once, but base by base, almost like a zipper with one link at a time,” Cunningham said. “The cool thing is that when the protector is replaced by the target snRNA, the target is smaller than the protector, so when the protector is removed, the exposed, available nucleic acid bases can be captured by the biosensor that has been specifically designed with a complimentary sequence.”
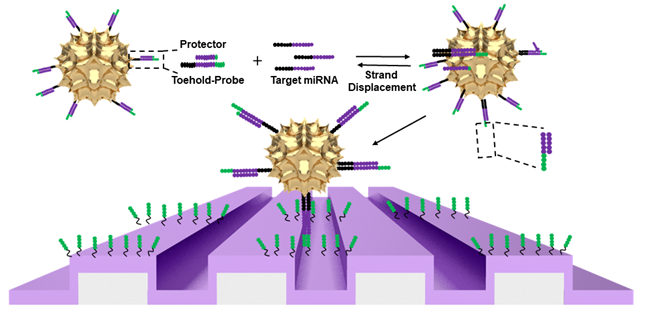
Figure of gold nanoparticles capturing target miRNA via Activate Capture + Digital Counting (AC+DC) process
Cunningham’s team named this biochemical process Activate Capture + Digital Counting, or AC+DC.
The target snRNAs, once captured by the AuNPs and the biosensor, are then counted using a red LED that is reflected into a simple, ordinary webcam. AuNPs are engineered to be very efficient absorbers of red light, so they appear as dark spots that researchers can count.
This testing method could easily be altered to diagnose other cancers or diseases, since there are RNAs associated with every kind of cancer that is thought to be involved in controlling gene expression and used by cancer cells to evade processes of restricting proliferation and metastasis.
The PRAM AC+DC technique may also have applications in monitoring the effectiveness of cancer therapies, where a baseline could be established to determine whether a therapy is having a positive or negative effect. This simple, low-cost blood test would be much less invasive than a biopsy, allowing for close monitoring of treatment efficacy and signs of rejection in patients.
– Written by the CCIL Communications Team
Brian Cunningham is a Cancer Center at Illinois program leader for the Cancer Measurement Technology program and a professor of electrical and computer engineering. Cunningham is also affiliated with the department of bioengineering and is the Director of the Micro and Nanotechnology Laboratory.
This work was supported by the National Institutes of Health (NIH), Carl R. Woese Institute for Genomic Biology, and Nick Holonyak Jr. Micro and Nanotechnology Laboratory.
The paper “Digital-resolution and highly sensitive detection of multiple exosomal small RNAs by DNA toehold probe-based photonic resonator absorption microscopy” is available online. DOI: 10.1016/j.talanta.2022.123256

